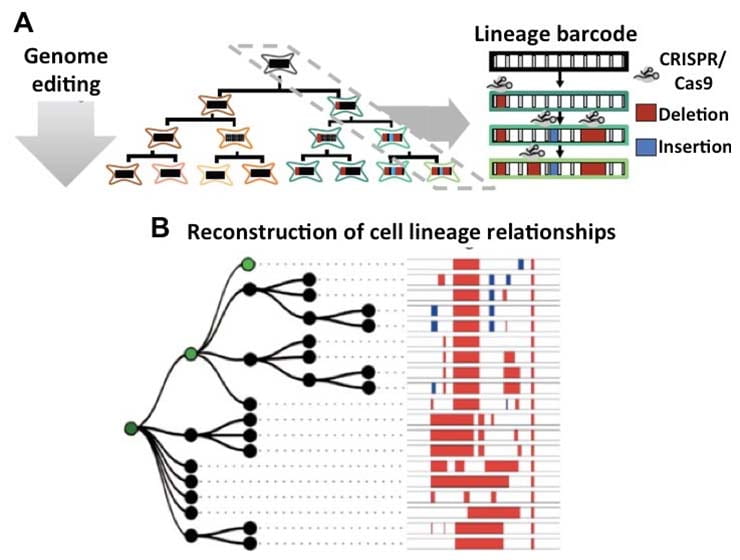
Although plant have long been used for analyses of cell fate, these studies have historically utilized X-irradiation, transposon activation, or transgenic markers to generate a relatively small number of readily detectable, genetically mosaic cells early in ontogeny, whose cell-lineage and cell fate is then inferred at later time-points in development. While powerful, previous studies have been hampered by the inability to track the lineages of all cells, tissues, and organs in a single experiment. New genomic technologies enable whole-organism lineage tracing (McKenna et al., 2016) that can be applied to answer long-standing questions in shoot ontogeny. In addition to lineage analyses of entire organisms as illustrated above, we will use laser-microdissection RNA-seq of amplified, edited barcode DNA to analyze patterns of shared and unique GESTALT barcode edits on cells microdissected from known locations in developing embryos and shoots, to investigate the cell lineage relationships among distinct cells, tissues and organs. Furthermore, because barcode edits can be sampled from genomic DNA or mRNA, GESTALT technology will be used in combination with scRNA-seq to investigate the relationship between cell lineage and cell-specific gene expression in maize shoots. Thus, this AIM will enable investigations of longstanding paradigms and hypotheses in plant cell lineage.


