A collaborative project with the PI Jianming Yu (Iowa State U), Bing Yang (U. Missouri), and Mark Lubkowitz (St. Michael’s College)
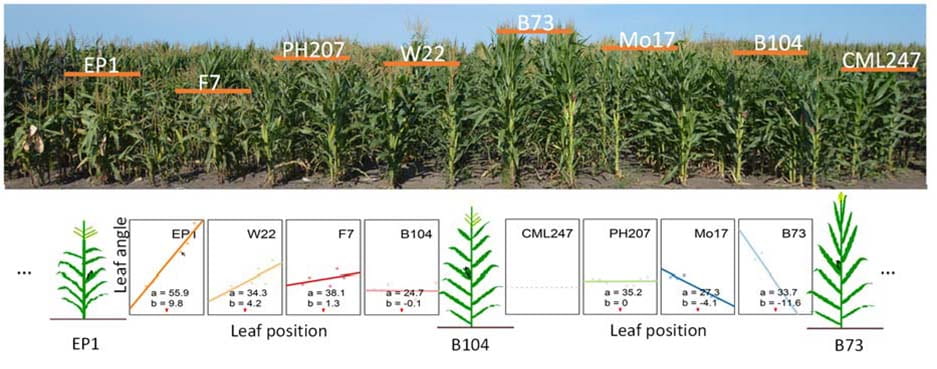
Leaf angle is typically measured on a single leaf per plant (e.g., at one leaf above the ear node, or at the flag leaf) in large-scale genetic studies. However, leaf angle variation across the canopy (i.e., variation among different leaves of the same plant) has not been well studied, even though diverse inbred lines are found to be extremely polymorphic for this “within-plant leaf angle variation” phenotype. Our long-term goal is to enrich fundamental understanding of the genetic control of leaf angle variation across the canopy in maize, and to provide mechanistic insights into genetic manipulation of plant architecture for continued crop improvement. Toward this end, we will use Laser-Microdissection RNA-sequencing to identify of genes underlying leaf angle variation across the canopy and investigate blade/sheath boundary-specific expression patterns in CRISPR-generated leaf angle mutants.


