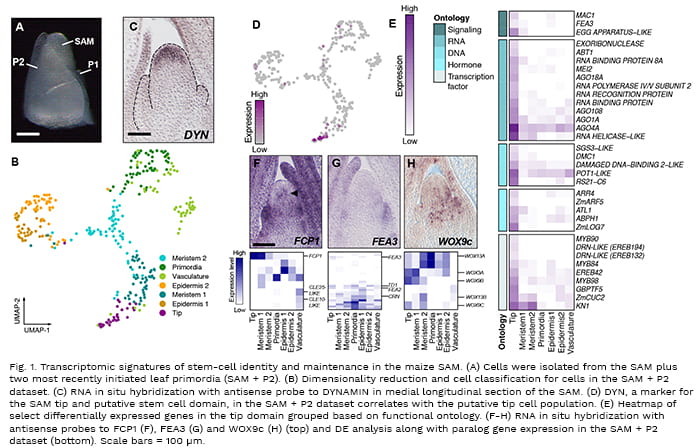
Single-Cell Transcriptomic Analyses of Shoot Meristem Ontogeny and Function
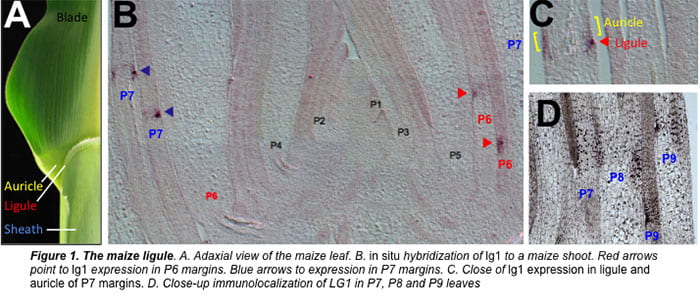
Ligule & Auricle Development in the Proximal-Distal Axis of the Maize Leaf
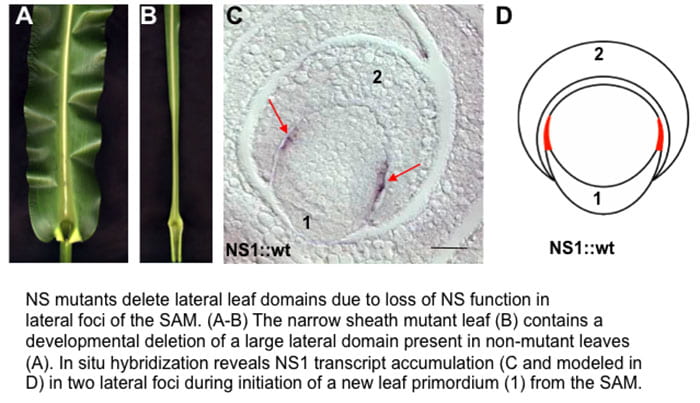
Comparative Analyses of planar growth in plant organs: WOX3 function
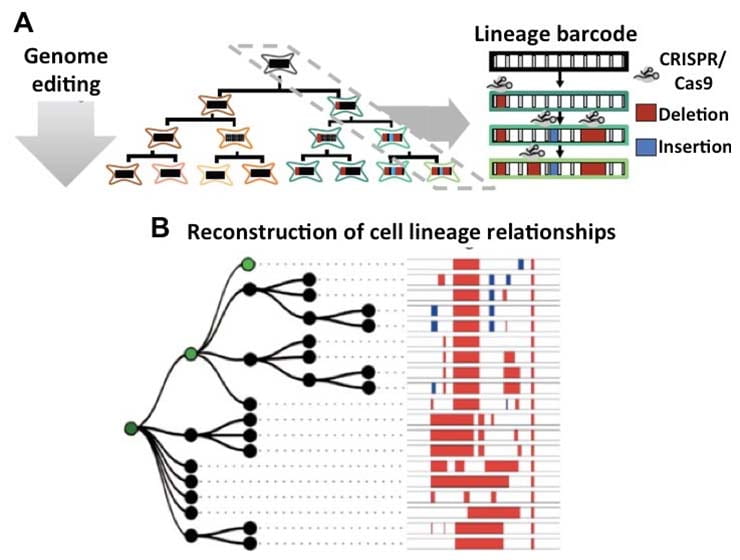
Whole Plant Cell Lineage Analyses: deciphering global, cell-fate acquisition
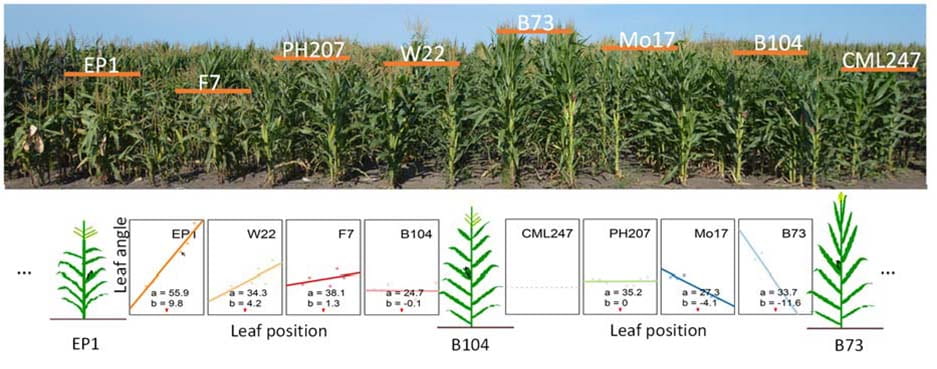
Genome-wide Dissection of Leaf Angle Variation Across the Canopy in Maize